L1Scouting analysis on SWAN with coffea + dask distributed
- Contact: Rocco Ardino (rocco.ardino\@cern.ch)
- Reference: https://swan.docs.cern.ch/condor/intro/
- Disclaimer: the status of this guide is still unstable, in particular for the choice of the software stack in SWAN
Configure SWAN
Open SWAN session: * https://swan-k8s.cern.ch/
Set the following configuration: * Tick "Try the new JupyterLab interface (experimental)" * Software stack: "Bleeding Edge" (it will probably need to be changed in the near future) * Memory: 16 GB * HTCondor pool: "CERN HTCondor pool"

When on jupyter lab interface: * Open terminal and run
kinit
~/.config/dask/jobqueue-cern.yaml (changing the user name and/or uncommenting the already existing config yaml file):
jobqueue:
cern:
name: dask-worker
# Default job resource requests
cores: 1
memory: "8 GiB"
log-directory: "/eos/user/r/rardino/dask_logs"
myschedd bump
- open on the left toolbar the dask section
- create a new cluster, select adaptive scaling, choose the amount of workers you need (don't allocate too much otherwise the condor jobs containing the worker script will get very low priority)
- wait for the worker jobs to be submitted
- open a notebook and connect the cluster (in the cluster panel on the lower left after clicking the dask symbol in the toolbar, just click on
<>)
Example notebook with Coffea + Dask for $m_{\mu\mu}$ invariant mass with scouting data
Packages
import os
import hist
import dask
import awkward as ak
import hist.dask as hda
import dask_awkward as dak
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
import mplhep as hep
from matplotlib.ticker import FuncFormatter
import uproot
# coffea packages needed
from coffea.nanoevents import NanoEventsFactory, BaseSchema
from coffea.nanoevents.methods import candidate
from coffea import processor
from coffea.dataset_tools import (
apply_to_fileset,
max_chunks,
preprocess,
)
# dask client
from dask.distributed import Client
client = Client("tls://10.100.165.46:30895")
Few configurations
# some configurations
hep.style.use(hep.style.ROOT)
hep.style.use(hep.style.CMS)
rlabel = "Level-1 trigger scouting 2024 (13.6 TeV)"
petroff_10 = ["#3f90da", "#ffa90e", "#bd1f01", "#94a4a2", "#832db6", "#a96b59", "#e76300", "#b9ac70", "#717581", "#92dadd"]
plt.rcParams["axes.prop_cycle"] = plt.cycler('color', petroff_10)
# example fileset (change number of lumisections if you don't have a lot of workers)
run_num = 383996
fill_num = 9969
nfiles = 10 # change to -1 if you want all the data set from that run
# example fileset (change number of lumisections if you don't have a lot of workers)
datapath = f"/eos/cms/store/cmst3/group/daql1scout/run3/ntuples/zb/run{run_num}/"
fileset = {
f'L1ScoutingZB-run{run_num}': {
"files": {
f"file:{datapath}/{f}" : "Events" for f in os.listdir(datapath)[:nfiles]
}
},
}
fileset_lsblk = {
f'L1ScoutingZB-run{run_num}': {
"files": {
f"file:{datapath}/{f}" : "LuminosityBlocks" for f in os.listdir(datapath)[:nfiles]
}
},
}
nOrbits = 0
for f in fileset_lsblk[f"L1ScoutingZB-run{run_num}"]["files"].keys():
nOrbits += uproot.open(f)["LuminosityBlocks"]["nOrbits"].array()[0]
print("Total number of orbits:", nOrbits)
Example Coffea processor for dimuon analysis
class MyProcessor(processor.ProcessorABC):
def __init__(self):
pass
def process(self, events):
dataset = events.metadata['dataset']
muons = ak.zip(
{
"pt" : events["L1Mu_pt"],
"eta" : events["L1Mu_etaAtVtx"],
"phi" : events["L1Mu_phiAtVtx"],
"mass" : dak.zeros_like(events["L1Mu_pt"]) + 0.1057,
"charge" : events["L1Mu_hwCharge"],
},
with_name="PtEtaPhiMCandidate",
behavior=candidate.behavior,
)
dataset_axis = hist.axis.StrCategory([], growth=True, name="dataset", label="Primary dataset")
mass_axis = hist.axis.Regular(200, 0, 200, flow=None, name="mass", label=r"$m_{\mu\mu}$ (GeV)")
h_mumu_ss = hda.hist.Hist(dataset_axis, mass_axis, storage="weight", label="SS")
h_mumu_os = hda.hist.Hist(dataset_axis, mass_axis, storage="weight", label="OS")
cut_kin = (muons.pt > 8) & (np.abs(muons.eta) < 0.83)
cut = (ak.num(muons[cut_kin]) == 2) & (ak.sum(muons[cut_kin].charge, axis=1) == 0)
dimuon = muons[cut][:, 0] + muons[cut][:, 1]
h_mumu_os.fill(dataset=dataset, mass=dimuon.mass)
cut = (ak.num(muons[cut_kin]) == 2) & (ak.sum(muons[cut_kin].charge, axis=1) != 0)
dimuon = muons[cut][:, 0] + muons[cut][:, 1]
h_mumu_ss.fill(dataset=dataset, mass=dimuon.mass)
return {
dataset: {
"entries": ak.num(events, axis=0),
"mmumu_ss": h_mumu_ss,
"mmumu_os": h_mumu_os,
}
}
def postprocess(self, accumulator):
pass
Run the machinery
dataset_runnable, dataset_updated = preprocess(
fileset,
align_clusters=False,
step_size=10_000_000,
files_per_batch=1,
skip_bad_files=True,
save_form=False,
)
to_compute = apply_to_fileset(
MyProcessor(),
max_chunks(dataset_runnable, 300),
schemaclass=BaseSchema,
)
(out,) = dask.compute(to_compute)
print(out)
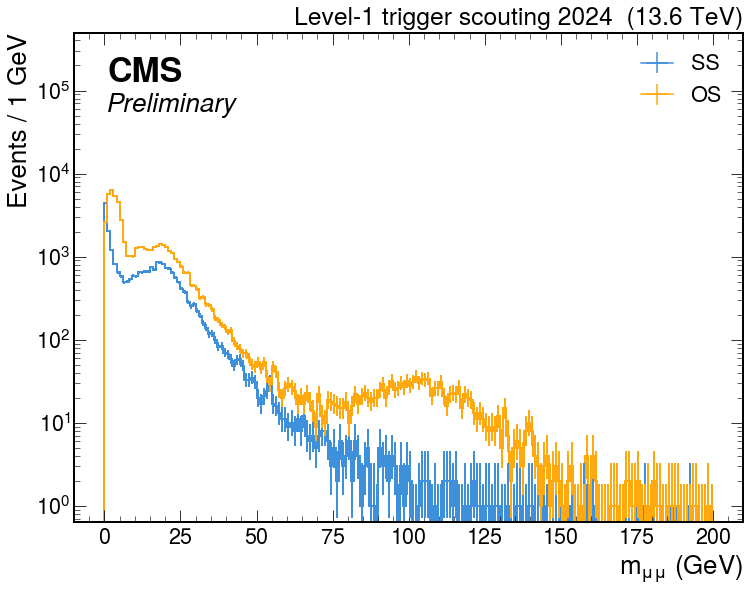
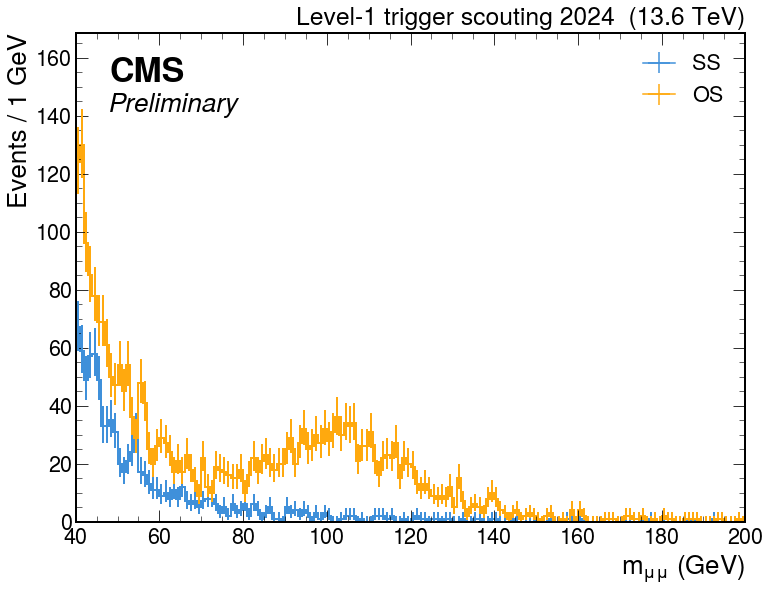
Plot
hep.style.use(hep.style.ROOT)
hep.style.use(hep.style.CMS)
fig, ax = plt.subplots(figsize=(12, 9))
hep.cms.label("Preliminary", loc=2, data=True, year=2024, rlabel=rlabel)
out["L1Scouting-run380346"]["L1Scouting-run380346"]["mmumu_ss"].plot(ax=ax, lw=2, label="SS")
out["L1Scouting-run380346"]["L1Scouting-run380346"]["mmumu_os"].plot(ax=ax, lw=2, label="OS")
plt.yscale("log")
xmin, xmax, ymin, ymax = plt.axis()
plt.ylim(ymin, ymax*50)
plt.xlabel("$m_{\mu\mu}$ (GeV)")
plt.ylabel("Events / 1 GeV")
plt.legend()
plt.show()
Notes
- You can monitor the execution through the dask toolbar functionalities, like CPU usage per worker and the Progress bars for the workload
- Performance:
- 614 Lumisections
- Dataset with prescale 25 (1 orbit~event every 25)
- 100 workers of 8 GB each
- Coffea preprocessor
step_size=10_000_000 - Coffea preprocessor
files_per_batch=1 - Execution time: ~300 sec
- results:
Standalone example using uproot
# configuration and file list
step_size = 10_000_000
flist = fileset[f"L1ScoutingZB-run{run_num}"]["files"]
flist = [f + ":Events" for f in flist]
# declare histograms to fill
dataset_axis = hist.axis.StrCategory([], growth=True, name="dataset", label="Primary dataset")
mass_axis = hist.axis.Regular(200, 0, 200, flow=None, name="mass", label=r"$m_{\mu\mu}$ (GeV)")
h_mumu_ss = hda.hist.Hist(dataset_axis, mass_axis, storage="weight", label="SS")
h_mumu_os = hda.hist.Hist(dataset_axis, mass_axis, storage="weight", label="OS")
for i,events in enumerate(uproot.iterate(files=flist, step_size=step_size, filter_name="/(bunchCrossing|nL1Mu|L1Mu_*)/")):
print(f"Batch {i+1}")
bxData = ak.zip(
{
"bx" : events["bunchCrossing"],
"nL1BMTFStub" : events["nL1BMTFStub"],
}
)
# declare sums
muons = ak.zip(
{
"pt" : events["L1Mu_pt"],
"eta" : events["L1Mu_etaAtVtx"],
"phi" : events["L1Mu_phiAtVtx"],
"mass" : ak.zeros_like(events["L1Mu_pt"]) + 0.1057,
"charge" : events["L1Mu_hwCharge"],
},
with_name="PtEtaPhiMCandidate",
behavior=candidate.behavior,
)
cut_kin = (muons.pt > 8) & (np.abs(muons.eta) < 0.83)
cut = (ak.num(muons[cut_kin]) == 2) & (ak.sum(muons[cut_kin].charge, axis=1) == 0)
dimuon = muons[cut][:, 0] + muons[cut][:, 1]
h_mumu_os.fill(dataset=dataset, mass=dimuon.mass)
cut = (ak.num(muons[cut_kin]) == 2) & (ak.sum(muons[cut_kin].charge, axis=1) != 0)
dimuon = muons[cut][:, 0] + muons[cut][:, 1]
h_mumu_ss.fill(dataset=dataset, mass=dimuon.mass)